i know, i know — it’s easier to spot inbreeding (or outbreeding) from the presence (or absence) of a lot of long runs of homozygosity (roh) in the genomes of individuals in a population rather than short roh (see for example the central/south and west asians in this post, populations which everyone knows are regular inbreeders), but i haven’t got any data on long roh for separate, sub-populations (like italians vs. europeans), so we’re gonna have to make do with short roh (for now). and anyway, even the amount of short roh is reduced via outbreeding (and increased via inbreeding), so you can use it as a tool to try to work out a population’s mating history. it’s just not as easy/obvious as with longer roh.
so … the map below is taken from Genomic and geographic distribution of SNP-defined runs of homozygosity in Europeans.
the samples come from:
– the rotterdam study – the netherlands
– popgen – northern germany – specifically the schleswig-holstein region (in deutsch if you like)
– the monica augsburg surveys – southern germany – from the city of augsberg and two neighboring counties
– and popres, which, since this is a study of europeans, i presume must mean that the samples came from both the lolipop study in london and the colaus study, lausanne, switzerland — i discussed those two studies in this previous post (scroll down).
again, the problem with taking samples from people living in big cities is that, even if they may be natives of whatever country they happen to live in, they, or some of their recent ancestors, may have migrated to the city — so, who knows, for instance, if the samples from rotterdam tell us anything about rotterdam or even the region of the country in which rotterdam is located. probably tells us something about the dutch, but even then….
these researchers — nothnagel et al. — chose to look at roh that were 1Mb in length. that’s shorter than the 1.5Mb roh as delineated by the researchers who looked at the roh in russian populations. also, nothnagel et al. weighted the average roh in each population according to how much linkage disequilibrium was (estimated to be) present in each population. don’t ask! no, really — don’t ask, because i don’t really understand why they did this. here’s the wikipedia page for linkage disequilibrium. i know that you can have more ld in an inbreeding population and — you guessed it! — less in an outbreeding one. and, of course, other things like bottlenecks can affect how much ld is present in a population. nothnagel et al. found different amounts of ld in the populations in this study and compensated for that, but again i’m not exactly sure why.
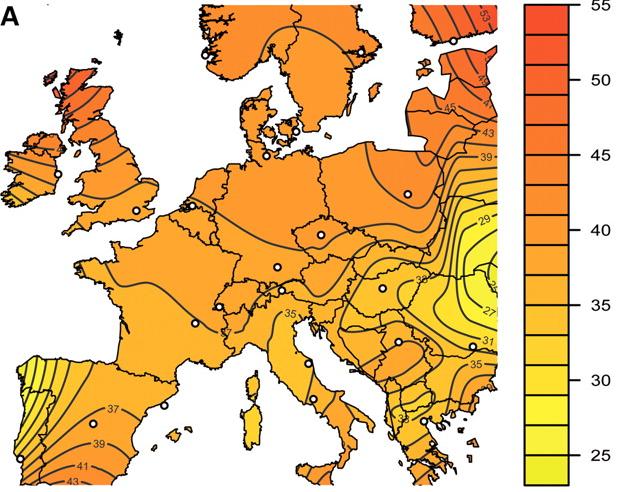
anyway … here’s what they found. this map shows the subpopulation averages of the weighted number of roh per individual (the contour lines are guesstimates — educated guesstimates, but still guesstimates):
if you look closely, you’ll see that there’s a sort-of central band of a relatively low average number of roh (between 37-39) that runs from southern england down through beligum/the netherlands (rotterdam) and northeast france, southern germany and switzerland. and, as the researchers observed, and as we saw in the previous post on russia, the numbers of roh increase going northwards and decrease going south. until you get to southern spain and southern italy, southern greece, and (probably) a central spot in the balkans there, all regions where the average number of roh increases again. the researchers suggest that, perhaps, migration from northern africa to the iberian peninsula (that’s the only region for which they offer a possible explanation for this anomaly) explains the longer roh there — presumably they’re thinking of a bottleneck. maybe. but perhaps it’s due to greater historic inbreeding in southern spain — and southern italy and greece and the balkans. some data showing longer roh would help us tell one way or the other.
the researchers, btw, acknowledge that the areas indicated as having very low amounts of roh — colored in the lightest shades of yellow — i.e. northwest spain and eastern europe — are probably artifacts of the interpolation method that they used. also, for all you scots out there (you know who you are! (^_^) ), while i do predict that the average numbers of roh in scotland ought to be higher there than in england, note that there was no data for scotland included in this study, so the shades of the contours up there are wild guesses as well.
i’m quite surprised by the very low levels of roh in romania, but remember that one has to read this map with the underlying north-south differences in numbers of roh in mind, so perhaps the roh in romania really indicates an inbreeding/outbreeding rate in romania that is more like that found in, say, france/germany. dunno. in any event, it’s very interesting.
now i want to compare the average number of roh in eastern europe with western europe. that’s going to be kinda hard to do since 1) the two studies used different roh lengths (1Mb vs. 1.5Mb), and 2) the numbers from this study have been weighted. still, i think we can get at something of a (very!) rough picture by taking the numbers from germany as our starting point and using them to calibrate the results from the two studies. we can do this, i think, since the samples from germany came from the same sources in both studies — the popgen study for northern germany and the monica study for southern germany.
in the russian study, the samples from northern and southern germany were combined, so we only have one number for germany — which was lower than all the results from eastern europe, typically much lower (see map from previous post). the number of roh in the polish sample, for instance, was more than twice that found for the germans. the average number of roh in russia (Rus_HGDP) was also twice that of the germans. czechs, latvians, estonians — all higher than the germans.
now if we work westwards from germany using the results from the study in this post — the english, the dutch (rotterdam), and the swiss are all in the same range as the southern germans, while the southern french have an even lower average number of roh — and the irish (in dublin) and the czechs are in the same range as the northern germans. so all of these populations — and even the spanish and italians — have fewer roh on average than eastern europeans. which is what i would’ve guessed given what we know about the historic mating patterns of europeans beginning in the early medieval period (see mating patterns in europe series below ↓ in left-hand column).
maybe there’s another explanation for this difference between western and eastern europe — and for the apparent differences between central and southern europe. like i said above, a study or two looking at longer roh would help to clear up the picture one way or the other.
previously: russians, eastern europeans, runs of homozygosity (roh), and inbreeding and ibd and historic mating patterns in europe and ibd rates for europe and the hajnal line and runs of homozygosity and inbreeding (and outbreeding) and runs of homozygosity again
(note: comments do not require an email. ruh roh!)
Long roh – clanism, short roh – nationalism?
It’s interesting that Finland (scoring high on this and the previous map) has as the only nordic country rejected multiculturalism. Perhaps the Sami admixture has inoculated them.
Finland seems to be an interesting culture, transitional between behaviors of inbreeders the behaviors to outbreeders, very much like Japan.
Perhaps there is an additional complication to the story…
“but i haven’t got any data on long roh for separate, sub-populations …”
Come to think of it, that’s going to require a lot of samples in a very large sample space. Tens of thousands, maybe hundreds of thousands of genomes even in a society as small as Italy. Or am I wrong about this? Bring in the mathematicians.
re: Finland: I’ve heard a few Finns claim that rural Finland is one of the most inbred parts of Europe. Maybe there’s something to that…? It would fit the hypothesis, given the history of the True Finns:
The predecessor of the True Finns was the Finnish Rural Party (Suomen maaseudun puolue, SMP), founded by Centre Party dissident Veikko Vennamo in 1959. Vennamo ran into serious disagreement particularly with Arvo Korsimo, then party secretary of the Agrarian League, and was excluded from the parliamentary group. As a result, Vennamo immediately started building his own organization and founded a new party, the Finnish Rural Party.
@staffan – “Long roh – clanism, short roh – nationalism?”
maybe!
@staffan – “It’s interesting that Finland (scoring high on this and the previous map) has as the only nordic country rejected multiculturalism.”
@jayman – “Finland seems to be an interesting culture, transitional between behaviors of inbreeders the behaviors to outbreeders, very much like Japan.”
@nydwracu – “re: Finland: I’ve heard a few Finns claim that rural Finland is one of the most inbred parts of Europe. Maybe there’s something to that…?”
so … what ya’ll are saying is that we need further research on the finns. … ok! (^_^)
@jayman – “Perhaps there is an additional complication to the story…”
there usually is. (~_^)
@luke – “Come to think of it, that’s going to require a lot of samples in a very large sample space. Tens of thousands, maybe hundreds of thousands of genomes even in a society as small as Italy. Or am I wrong about this?”
i dunno, but … yes, please!
and i’d also like LOTS of sequencing of genomes sampled from skeletal remains for historic populations, too, thankuverymuch.
(^_^)