in “Genomic Runs of Homozygosity Record Population History and Consanguinity” that i posted about yesterday, kirin, et. al., say:
“Europeans and East Asians have very similar ROH profiles in all but the shortest category (0.5-1 Mb). There are no significant differences between either the percentage of individuals with ROH of different lengths or sum length of ROH above different length thresholds (>1.5 Mb) for these two continental groupings (File S1). This is not surprising because both of these groups are mainly represented here by fairly large populations with no documented preference for consanguineous marriage.“
ehhhhhhh … well … if they’re talking about now, i.e. in the present, then yeah — that’s probably pretty right. but many of the european populations that they looked at (i.e. from the human genome diversity project [hgdp]), regularly practiced some to quite a lot of consanguineous marriages up until fairly recently. (i haven’t checked into the asian populations that they looked at.)
the european populations that they looked at are: the adygeis, the basques, french folks, italians, orcadians, russians, sardinians and tuscans.
the adygeis are the circassians and it’s my understanding that they have avoided cousin marriage for quite some time, although they are endogamous (obviously). the russians — religious russians, anyway — avoid first- and second-cousin marriage. but the basques and the french have had some signficant amounts of consanguineous marriage up until quite recently. and the italians and sardinians?! holy toledo! of all of these groups, it’s probably the tuscans that have avoided cousin marriage for the longest. (dunno about the orkney islanders.)
like i said yesterday, if anything, kirin, et. al., have probably got some of the most inbred europeans in their sample.
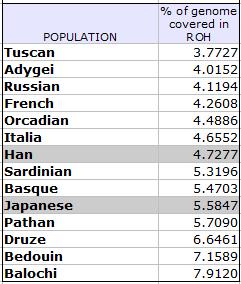
anyway … i took at look at their supplemental info [opens pdf] and found that they’ve included data for the proportion (percentage) of the genomes from each group that are covered in “runs of homozygosity” (roh). the more roh in your — or your population’s — genome(s), the more inbred you (all) are (or maybe the smaller your gene pool is — see yesterday’s post). when i took out just the europeans plus the han chinese and japanese and a couple of other interesting groups, here’s what i got:
most of the european groups have the least number of roh (these are roh of all different lengths). the han chinese are like the italians or the sardinians, who have a long and recent history of close marriages (not so much the northern italians) — and the japanese even more so. wikipedia tells us that cousin marriage was preferred in china until the mid-twentieth century, so there you go.
and the father’s brother’s daughter’s marriage groups? their roh are higher than the inbred europeans, the han chinese and the japanese.
you can see here, too, that the japanese have greater numbers of longer roh than french people (the black circles are the japanese, the orange circles are the french) — that means more recent inbreeding amongst the japanese (click on image for LARGER view – should open in new tab/window):
interestingly, many balochis (green circles) have fewer and shorter roh than the french — many have more and longer. dunno what that tells us about the balochi. new blood? tribes merging with (fairly) unrelated tribes? just plain ol’ out-marriage?
here are the percentages of the genomes covered by roh for each of the populations in the study in ascending order. i tried to match the colors for the continental groupings from the chart in yesterday’s post — dunno if i succeeded?:
previously: runs of homozygosity and inbreeding (and outbreeding)
(note: comments do not require an email. balochi farmer. (^_^) )


“dunno about the orkney islanders”
I was assuming a low base population would lead to a high ROH even if you avoided nearest cousins because everyone is pretty much a cousin just by the law of averages. Or maybe a low pop. trying to be as exogamous as possible under the circumstances has a different signature?
I don’t really understand the African pattern of lots of small ROH either.
@g.w. – “I was assuming a low base population would lead to a high ROH even if you avoided nearest cousins because everyone is pretty much a cousin just by the law of averages.”
yeah, that’s right. the thing with the orcadians, though, is that they’re a mutt group founded kinda recently. starting off with that mix of pictish, other scottish and viking genes will give you some low total roh rates, even if you inbreed. if they all consistently married their first cousins since the viking age, i think you’d see a lot of long roh in their genomes, but the “background” short roh count would be fairly low since they’re a mix of groups. in other words, the more family-related roh (the long roh) increases with close, recent inbreeding — the population-related roh (the short roh) increases with a bottleneck or decreases if you’ve got variety in your population
that’s why the africans have lots of short roh — they never went through any bottleneck. they have a lot of genetic variation at the population level. europeans & asians — a bottleneck a long time ago (out-of-africa or arabia or wherever); oceanians & native-americans — an even more recent bottleneck. that’s why they’ve got the most short roh.
(what i am not sure about is whether or not the orkney islanders are or were marrying their cousins much ever.)
So did the researchers of this study misrepresent their data? I was thinking that their sample sizes were very small.
@luke – “So did the researchers of this study misrepresent their data?”
no, that would be far too strong to say, i think.
the researchers said:
“This is not surprising because both of these groups are mainly represented here by fairly large populations with no documented preference for consanguineous marriage.”
if, by that, they mean now … today … right this moment as we’re
speakingtyping … then, yeah, that’s pretty right. most europeans and east asians (well, the chinese and japanese, anyway — i don’t know about the rest) are not marrying their cousins very much (consanguineous marriage=1st & 2nd cousin marriage).however, both groups that they looked at — east asians and mostly european populations on the periphery of europe — did practice “quite a bit” to “a lot” of cousin marriage up until quite recently (egs. mid-twentieth century for the chinese, 1960s for southern italians).
so, i don’t know what they mean by “no documented preference for consanguineous marriage.” if they mean today — yes, that’s right. but if one or two generations ago makes any difference to these long rohs (and i don’t know how much of a difference it would make), then they didn’t do their homework.
i think where the “misrepresentation” comes in (that’s still too strong of a word) — or “misunderstanding” comes in is that they seem to be assuming that southern italians, basques and russians are a good representation for all of europe. they’re not. there’s “core” europe and “peripheral” europe. these guys missed out on core europe (but it’s not their fault! ’cause there are no core european samples in the hgdp. edit: sorry. that’s not entirely right. there are french samples in the hgdp. but apart from those, all the rest are “peripheral” europeans. really peripheral if you count the circassians as europeans.).
@luke – “I was thinking that their sample sizes were very small.”
yeah, that must definitely be the case. but it’s early days in this sort of genetic research, isn’t it? — so you make do with what you got. (^_^) i look forward to when there are tens of THOUSANDS of genomes available out there and much more of this sort of research can be done. yipee! (^_^)